- 2026
-
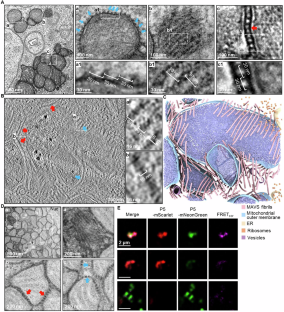
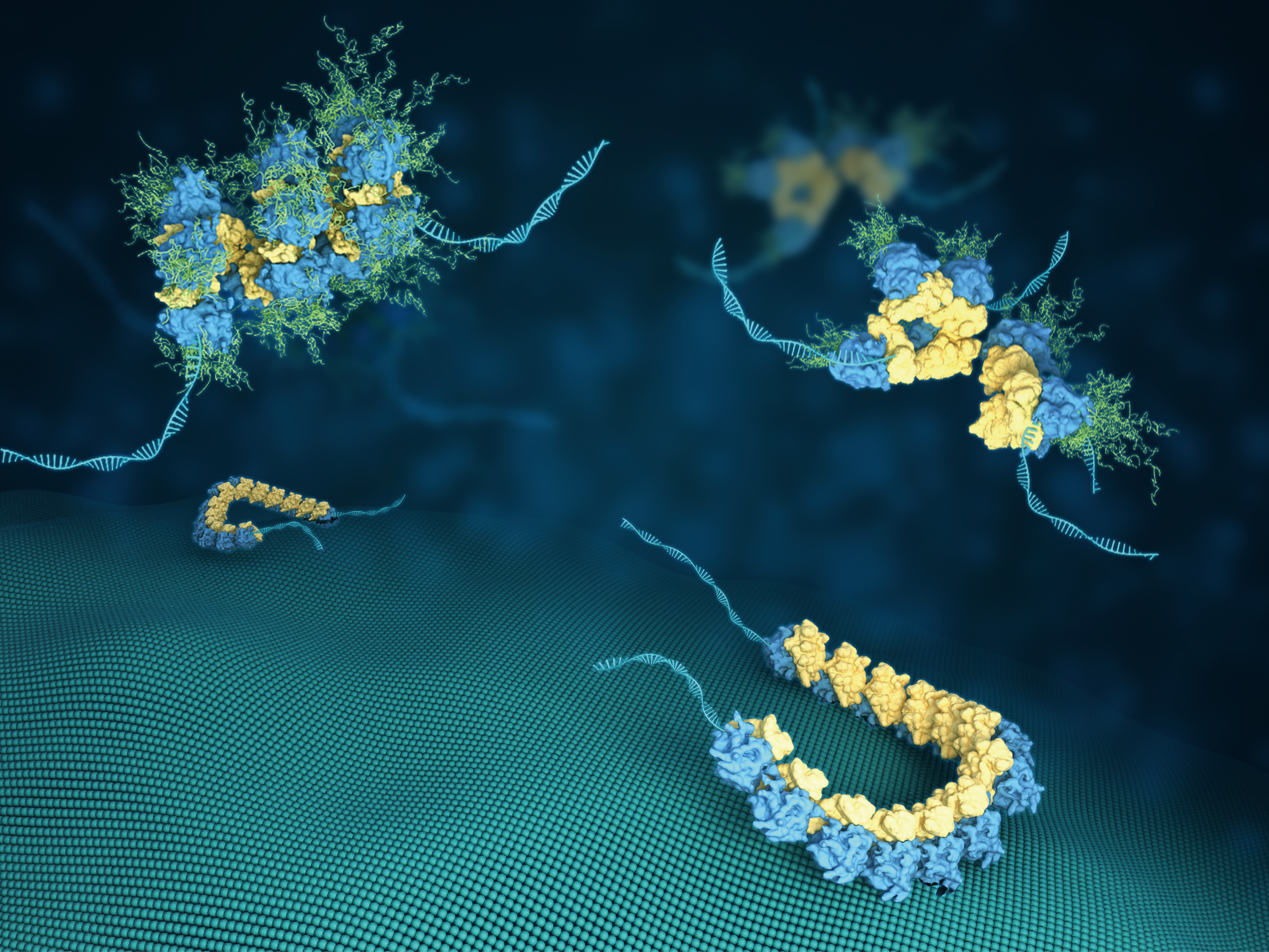
Yu, X., Wang P., Zhou N., Zhang L., Gong X., Yao L., Guo Q.#, Jiang Z.#, (2026). Prion-like MAVS fibrils stitch mitochondria to promote a rapid antiviral response. Cellular & Molecular Immunology 16, 9150.
- 2025
-
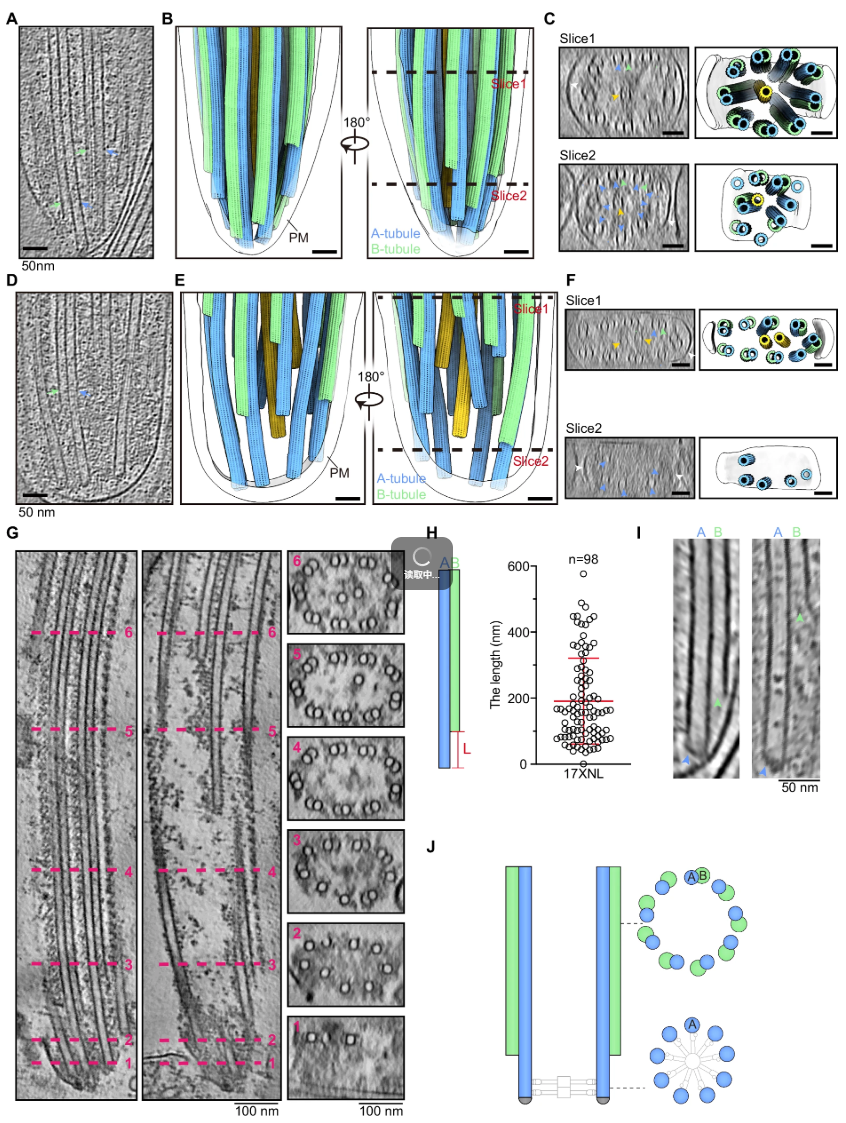
Yang, S., Ma S., Yuan C., Li Z., Ji F., Yao L., Cui H., Guo Q.#, Yuan J.#, (2025). A basal body microtubule singlet-to-doublet transition in Plasmodium male gametogenesis. Nature Communications 1672, 7681.
-
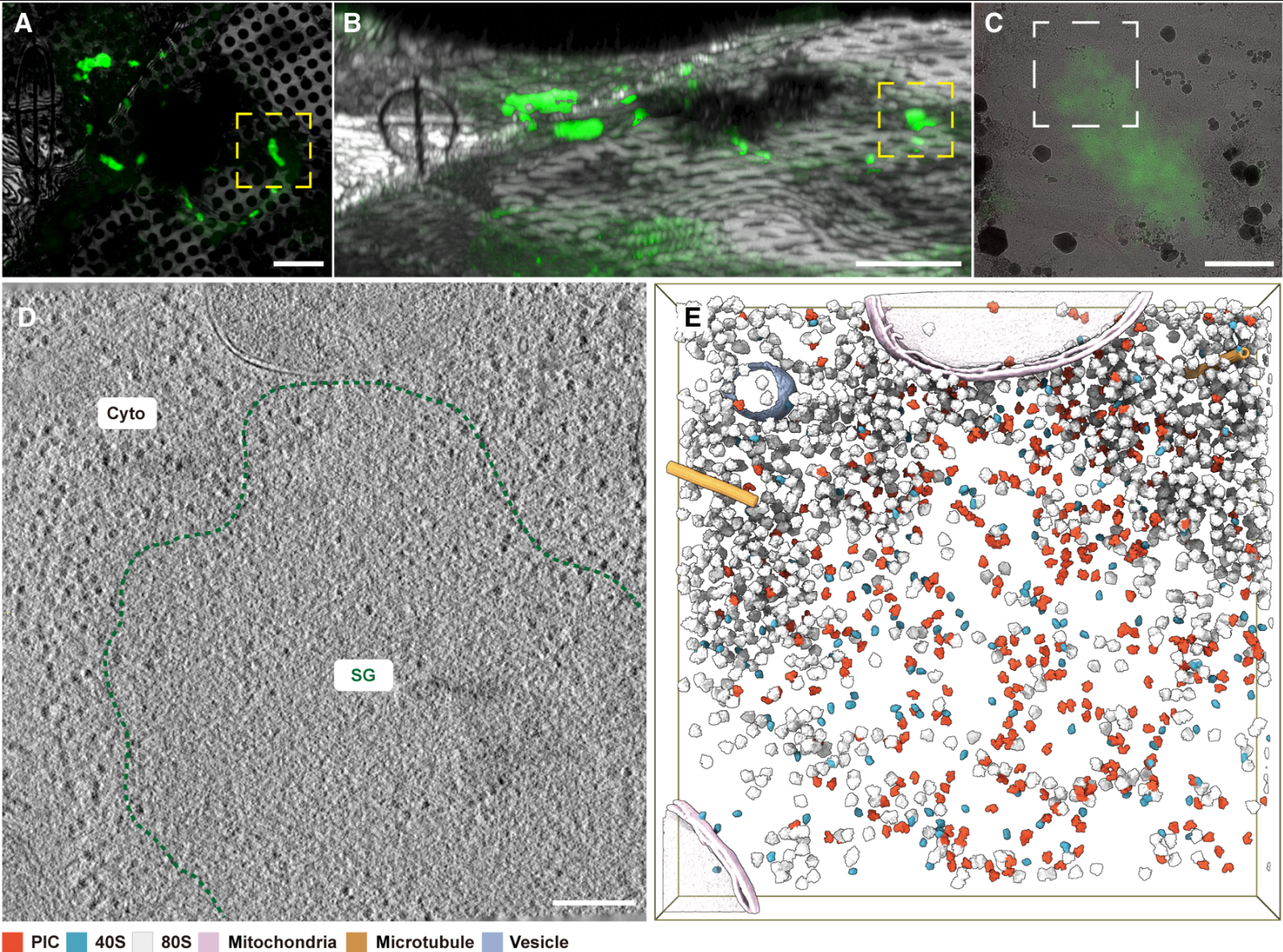
Wu, Y., Wang X., Meng L., Liao Z., Ji W., Zhang P., Lin J.#, Guo Q.#,(2025). Translation landscape of stress granules. Science Advances 11, eady6859
-
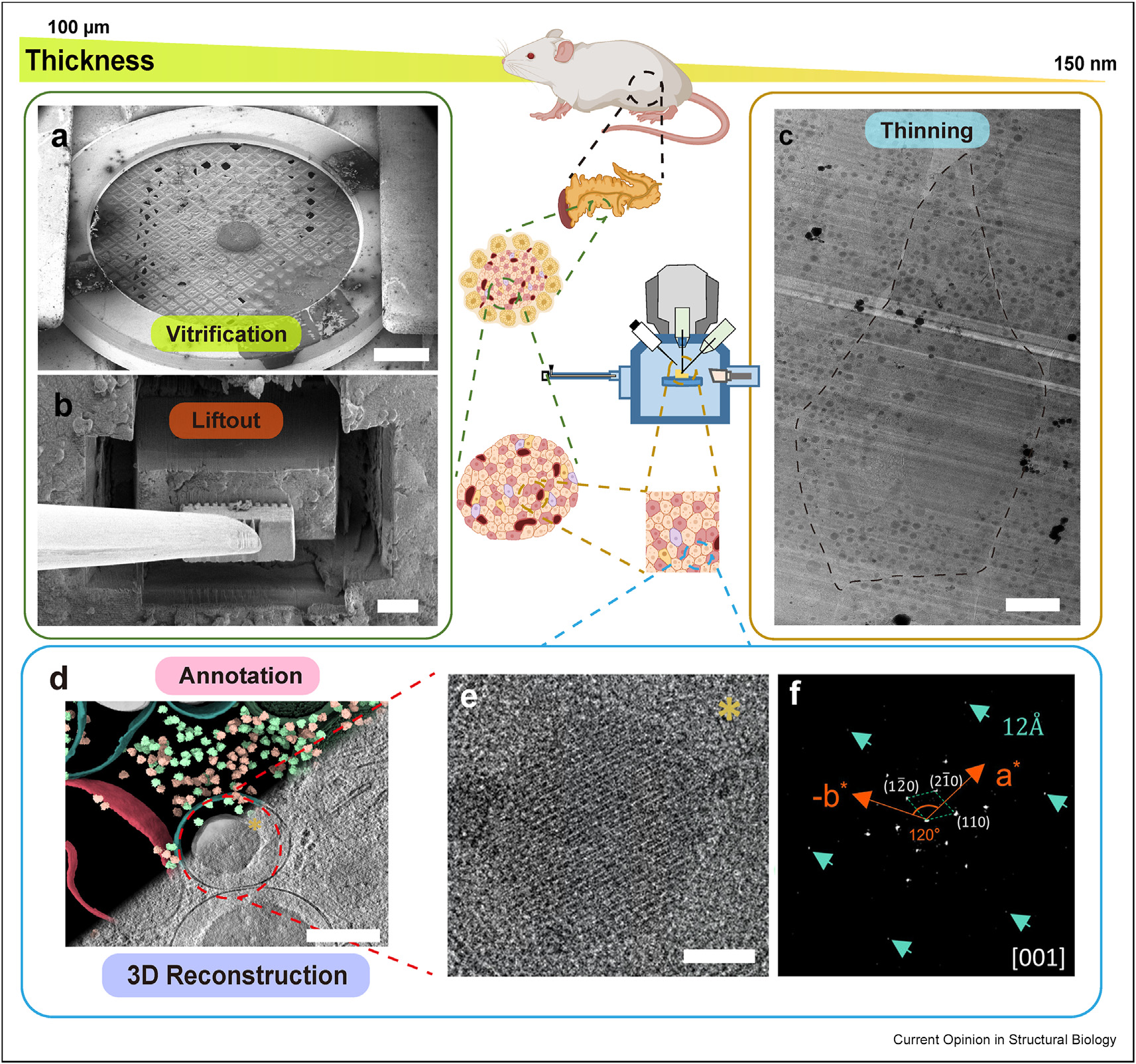
Chen, Z., Guo, Q.#, (2025). Innovations in cryo-electron tomography for tissues: Challenges and future prospects. Current Opinion in Structural Biology 93:103112
-
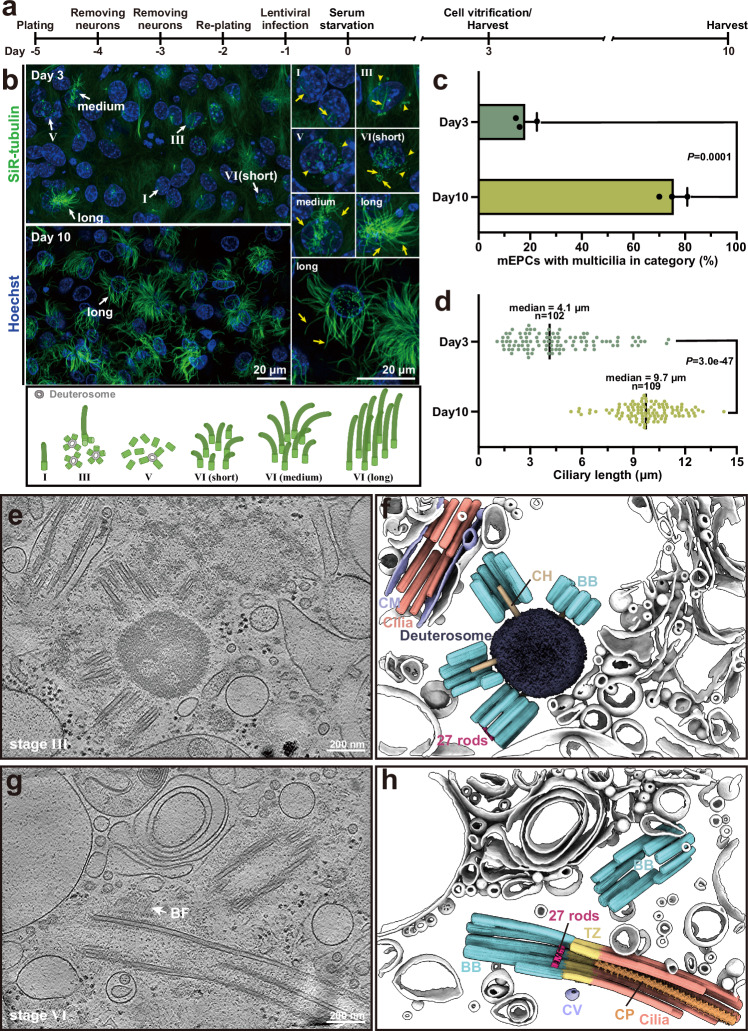
Ma, S., Li, L., Li, Z., Luo, S., Liu, Q., Du, W., Qiu, B., Gui, M.#, Zhu, X.#, & Guo, Q.# (2025). In situ cryo-electron tomography reveals the progressive biogenesis of basal bodies and cilia in mouse ependymal cells. Nature communications 16, 5932.
-

Luo, D., Zheng, L., Lu, M., Chen, Z., Wang, P., Guo, Q.#, Gao, N.#, (2025). Structural basis for prohibitin-mediated regulation of mitochondrialm-AAA protease. bioRxiv.
- 2024
-

Lu, M. *, Qian, Y., Ma, L., Guo, Q.#, and Gao, N.#, (2024). Molecular mechanism of the flotillin complex in membrane microdomain organization. bioRxiv.
-

Wu, L. *, Wang, J. *, Wang, Y. *, Yang, J., Yao, Y., Huang, D., Hu, Y., Xu, X., Wang, R., Du, W., Shi, Y., Li, Q., Liu, Lu., Zhu, Y., Wang, X., Guo, Q. #, Xu, L. #, Li, P. #, and Chen, X. #, (2024). CLCC1 Governs Bilayer Equilibration at the Endoplasmic Reticulum to Maintain Cellular and Systemic Lipid Homeostasis. bioRxiv.
-
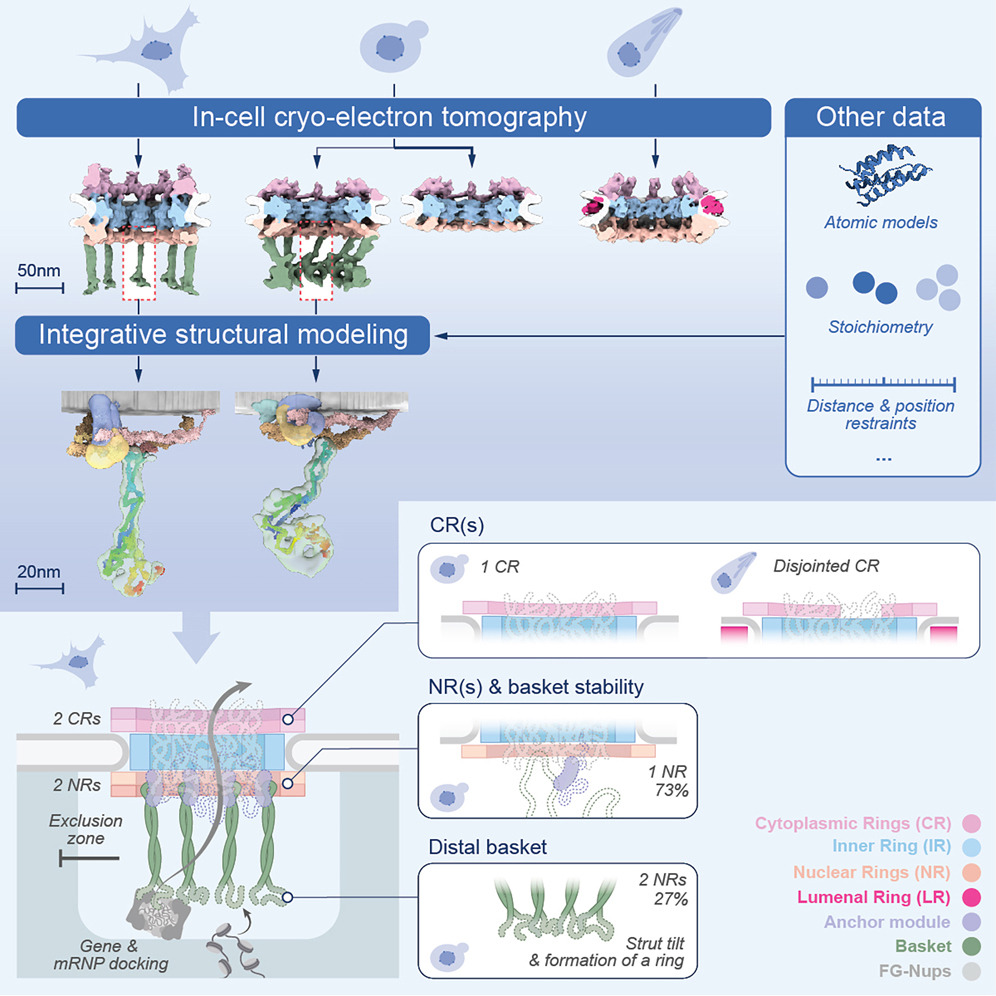
Singh, D.*, Soni, N.*, Hutchings, J.*, Echeverria, I., Shaikh, F., Duquette, M., Suslov, S., Li, Z., van Eeuwen, T., Molloy, K., Shi, Y., Wang, J., Guo, Q., Chait, B.T., Fernandez-Martinez, J., Rout, M.P.#, Sali, A.#, Villa, E.#, (2024). The molecular architecture of the nuclear basket. Cell 187, 5225-5227.
-
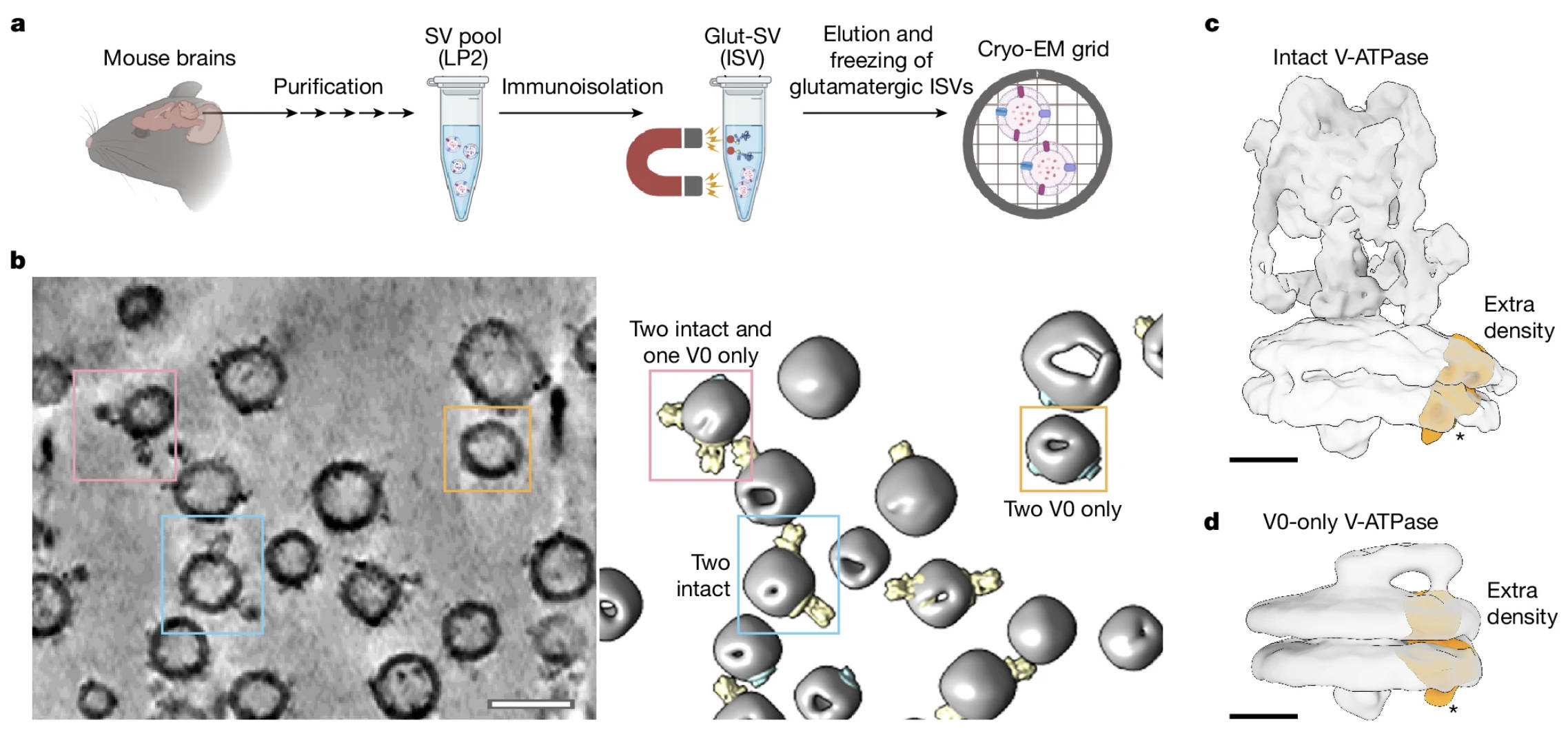
Wang, C.*, Jiang, W.*, Leitz, J.*, Yang, K., Esquivies, L., Wang, X., Shen, X., Held, R., Adams, D.J., Basta, T., Hampton, L., Jian, R., Jiang, L., Stowell, M.H.B., Baumeister, W., Guo, Q.#, Brunger, A.T.#, (2024). Structure and topography of the synaptic V-ATPase–synaptophysin complex. Nature 631, 899-904.
- 2023
-
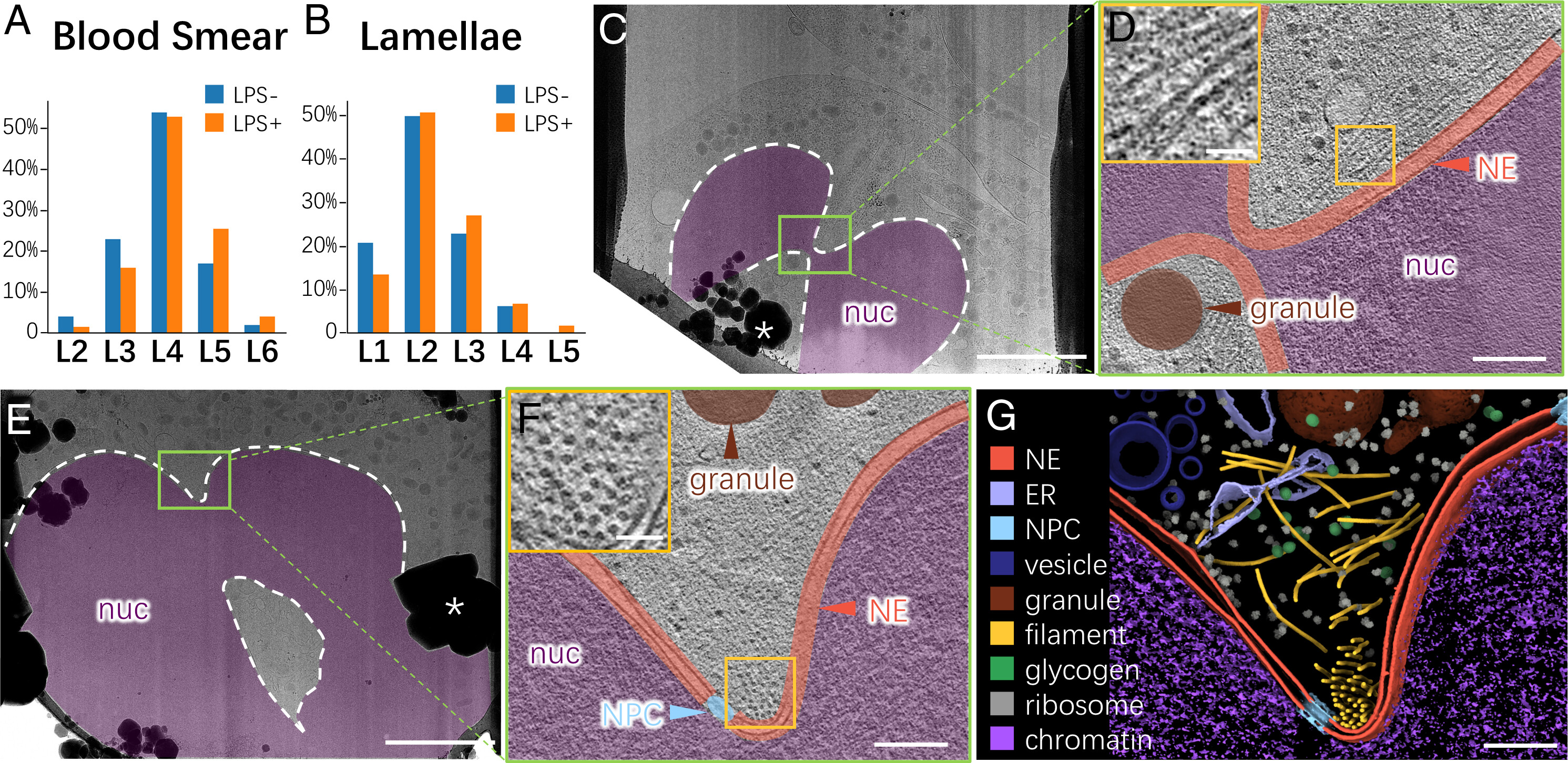
Liu, J.*, Li, Z.*, Li, M.*, Du, W., Baumeister, W., Yang, J.#, Guo, Q.#, (2023). Vimentin regulates nuclear segmentation in neutrophils. PNAS 120, e2307389120.
-
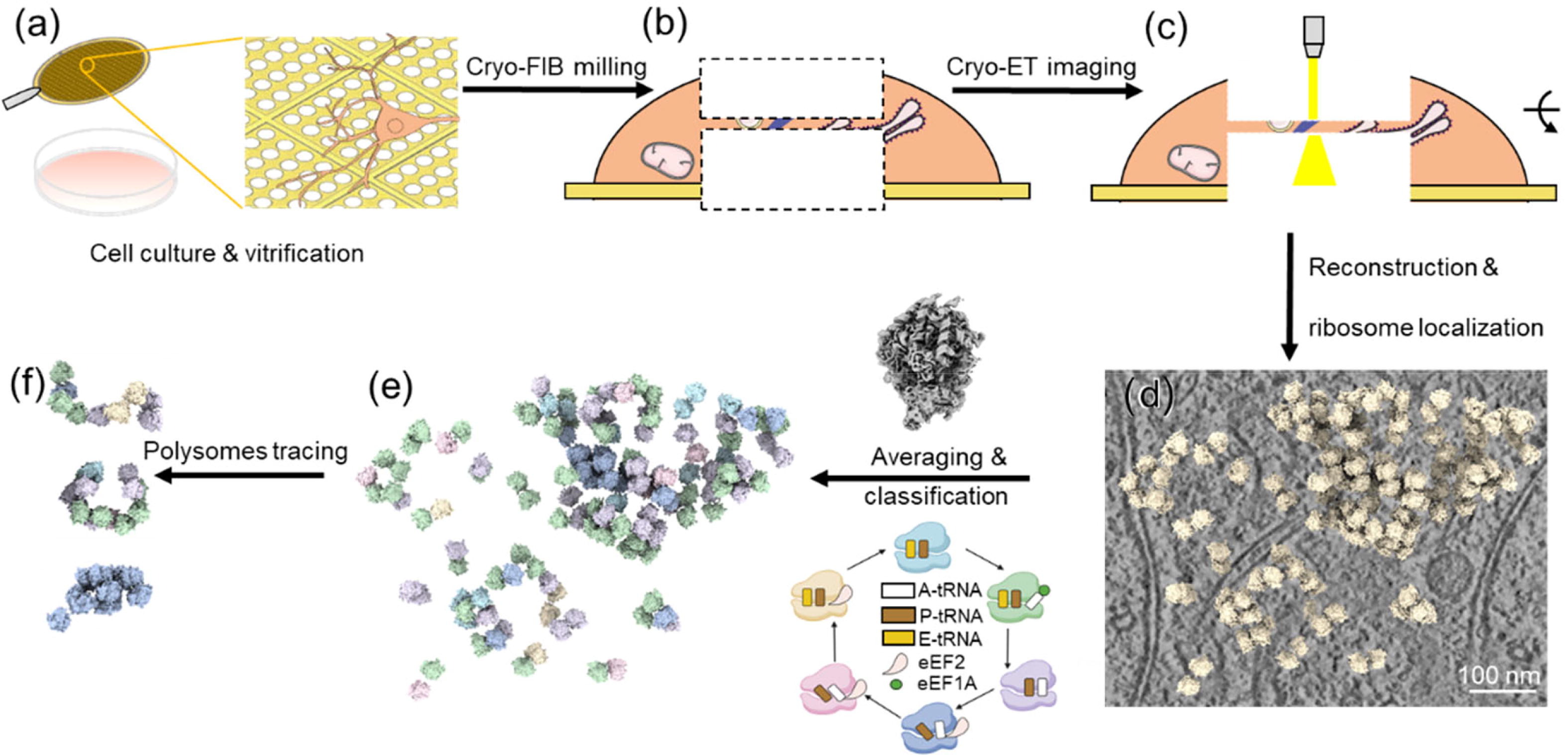
Guo, Q.#, Baumeister, W., Gao, N.#, (2023). Atomic structures of ribosomes at work captured by in situ cryo-electron tomography. Science Bulletin 68, 2671-2673.
-
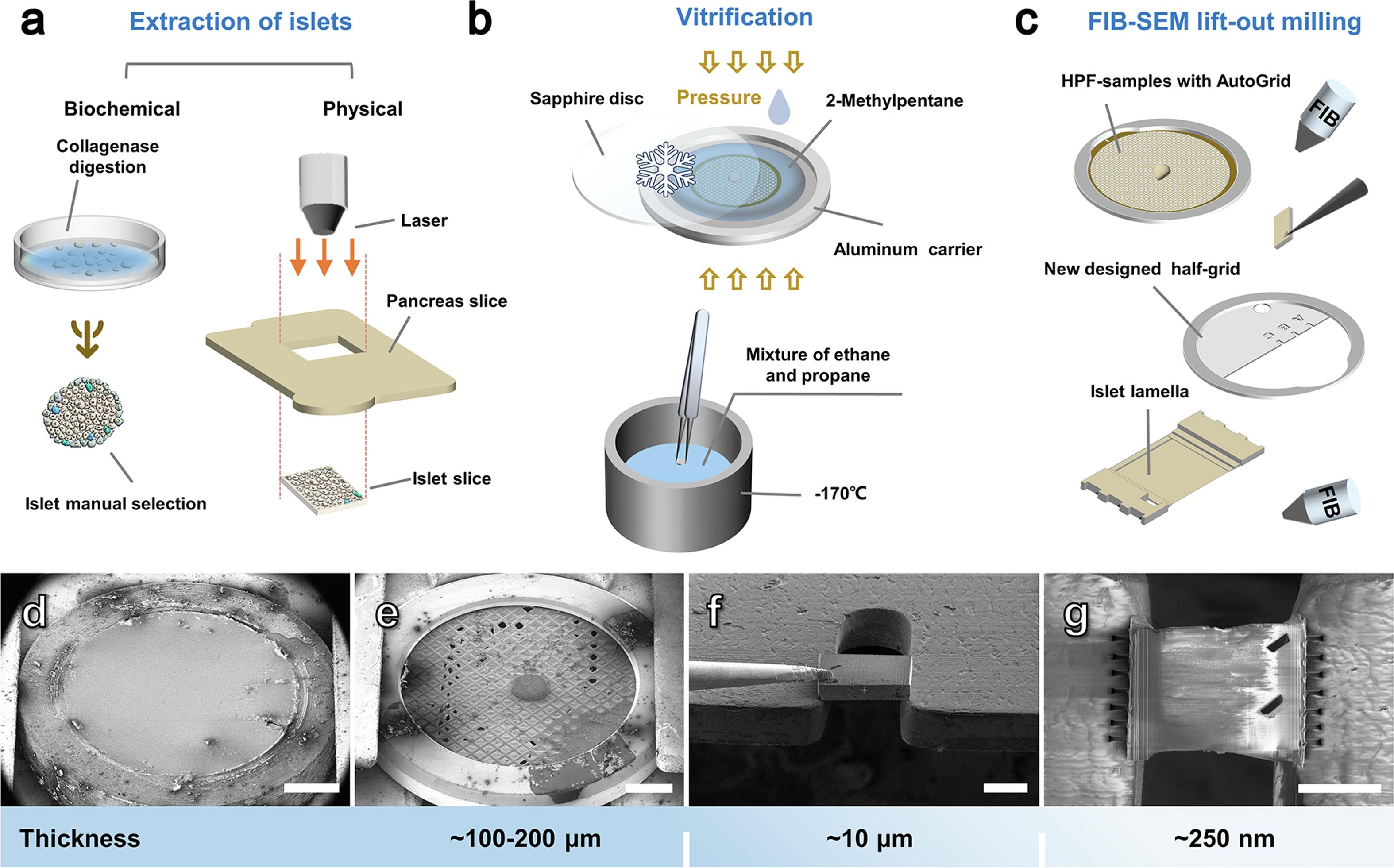
Wu, Y.*, Qin, C.*, Du, W.*, Guo, Z., Chen, L., Guo, Q.#, (2023). A practical multicellular sample preparation pipeline broadens the application of in situ cryo-electron tomography. Journal of Structural Biology 215, 107971. Cover Story.
-
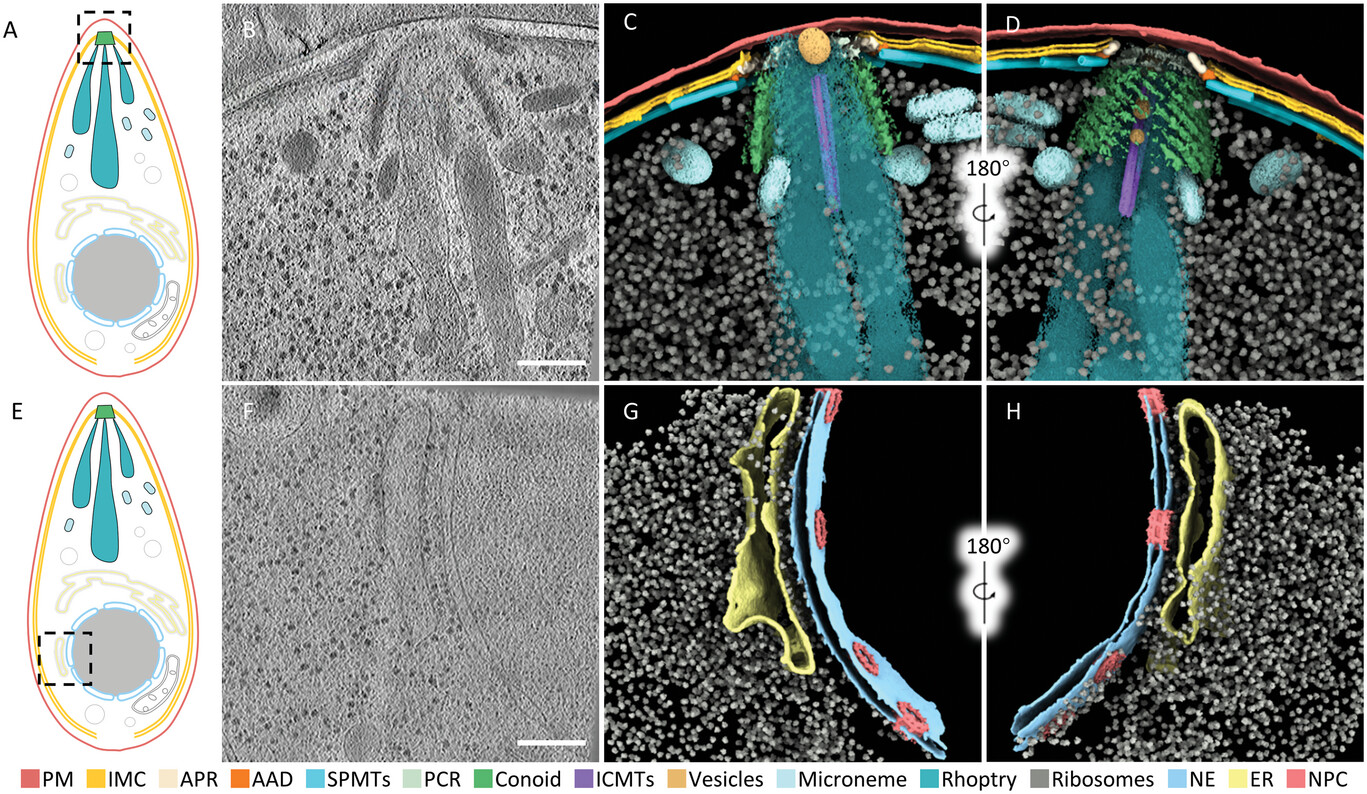
Li, Z., Du, W., Yang, J., Lai, D., Lun, Z., Guo, Q.#, (2023). Cryo-Electron Tomography of Toxoplasma gondii Indicates That the Conoid Fiber May Be Derived from Microtubules. Advanced Science 10, 2206595. Cover Story.
-
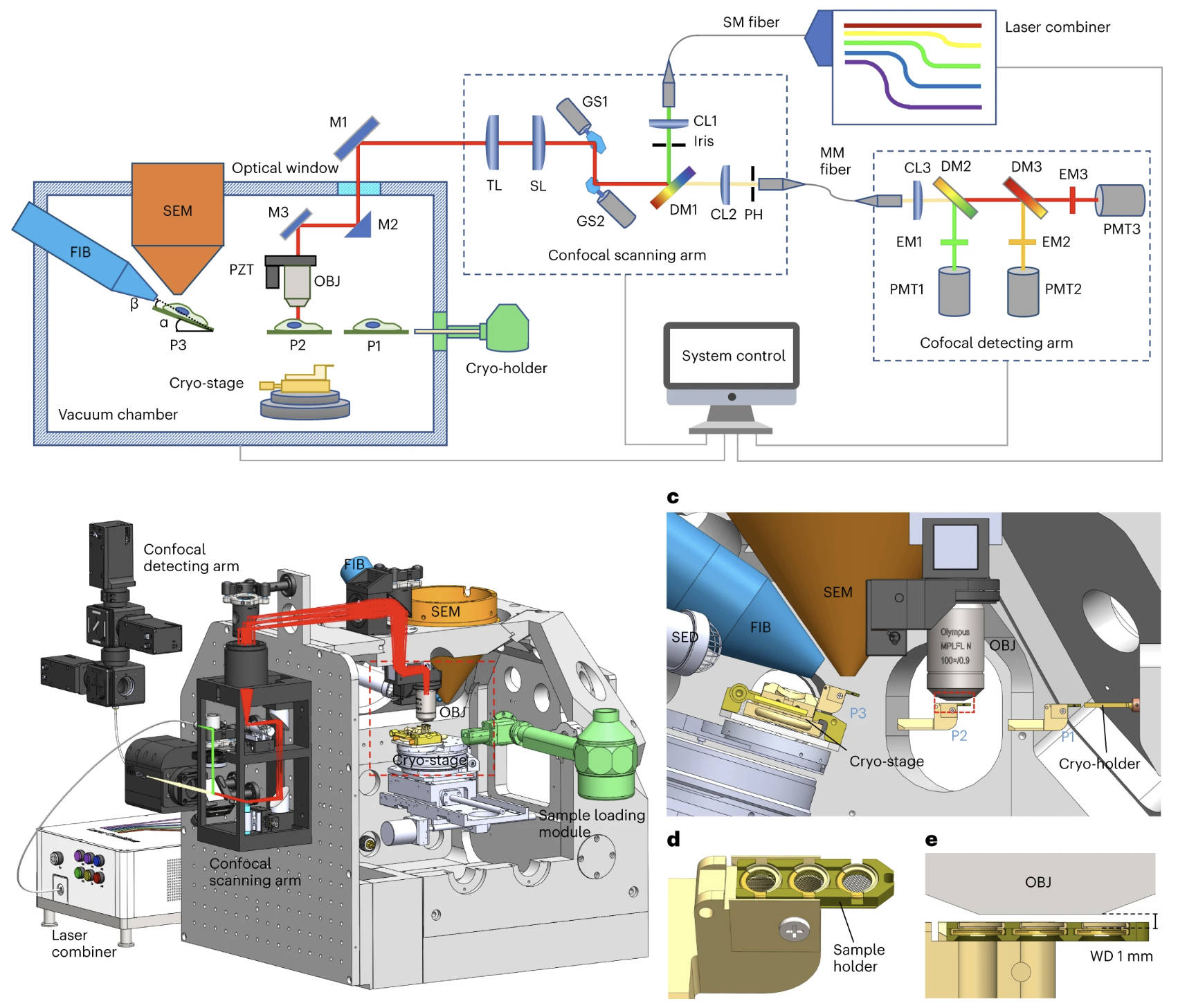
Li, W.*, Lu, J.*, Xiao, K.*, Zhou, M.*, Li, Y., Zhang, X., Li, Z., Gu, L., Xu, X., Guo, Q.#, Xu, T.#, Ji, W.#, (2023). Integrated multimodality microscope for accurate and efficient target-guided cryo-lamellae preparation. Nature Methods 20, 268-275.
- 2022
-
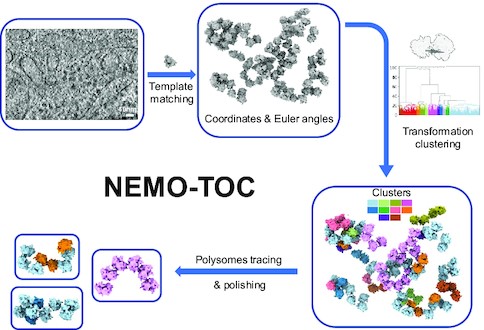
Jiang, W., Wagner, J., Du, W., Plitzko, J., Baumeister, W., Beck, F.#, Guo, Q.#, (2022). A transformation clustering algorithm and its application in polyribosomes structural profiling. Nucleic Acids Research 50, 9001-9011. Cover Story.
DOI: 10.1093/nar/gkac547
-
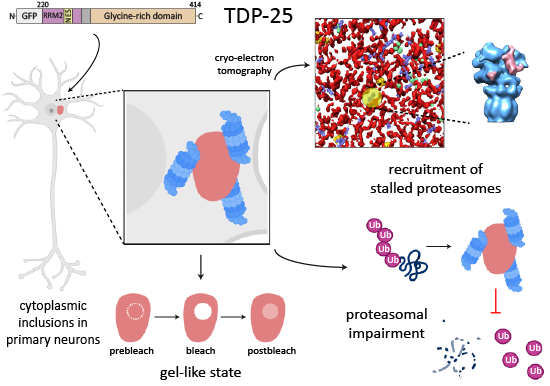
Riemenschneider, H., Guo, Q., Bader, J., Frottin, F., Farny, D., Kleinberger, G., Haass, C., Mann, M., Hartl, F.U., Baumeister, W., Hipp, M., Meissner, F., Fernández-Busnadiego, R.#, Edbauer, D.#, (2022) Gel-like inclusions of C-terminal fragments of TDP-43 sequester stalled proteasomes in neurons. EMBO Rep 23, e53890.
-
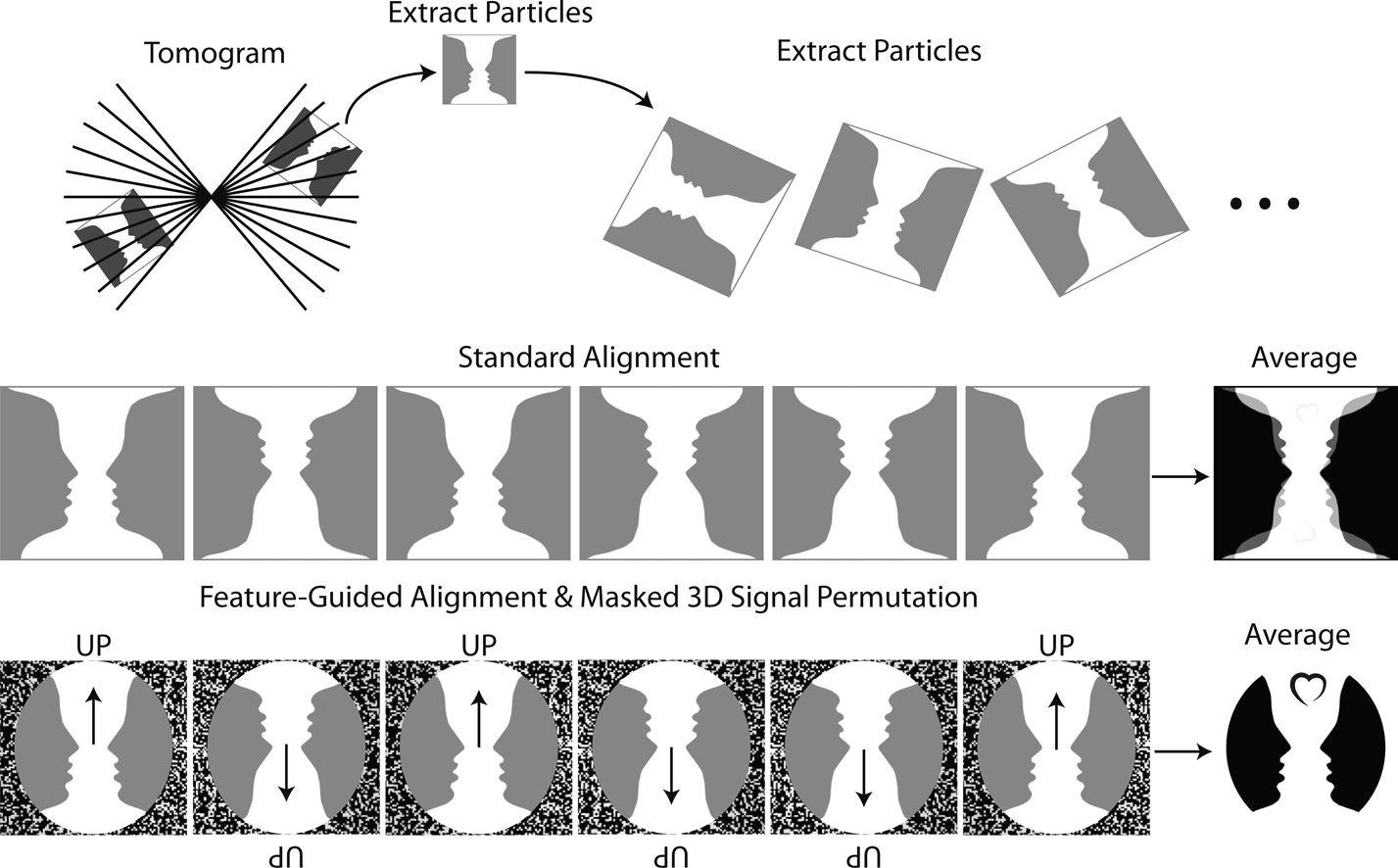
Peters, J.J., Leitz, J., Guo, Q., Beck, F., Baumeister, W. and Brunger, A.T.#, (2022) A feature-guided, focused 3D signal permutation method for subtomogram averaging. Journal of Structural Biology 214, 107851.
- 2021
-
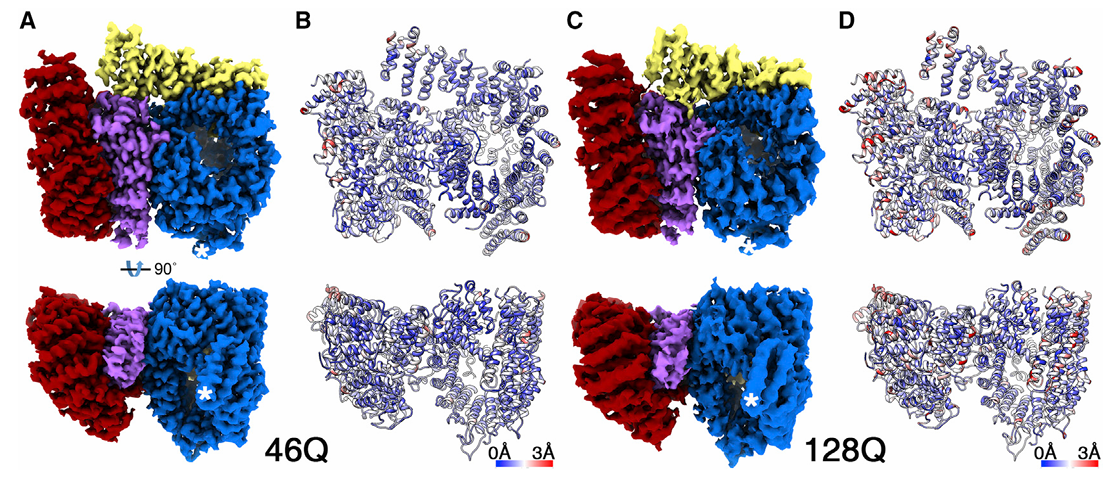
Huang, B.*, Guo, Q.*, Niedermeier, M.L., Cheng, J., Engler, T., Maurer, M., Pautsch, A., Baumeister, W., Stengel, F., Kochanek, S., et al. (2021). Pathological polyQ expansion does not alter the conformation of the Huntingtin-HAP40 complex. Structure 29, 804-809.e805.
-
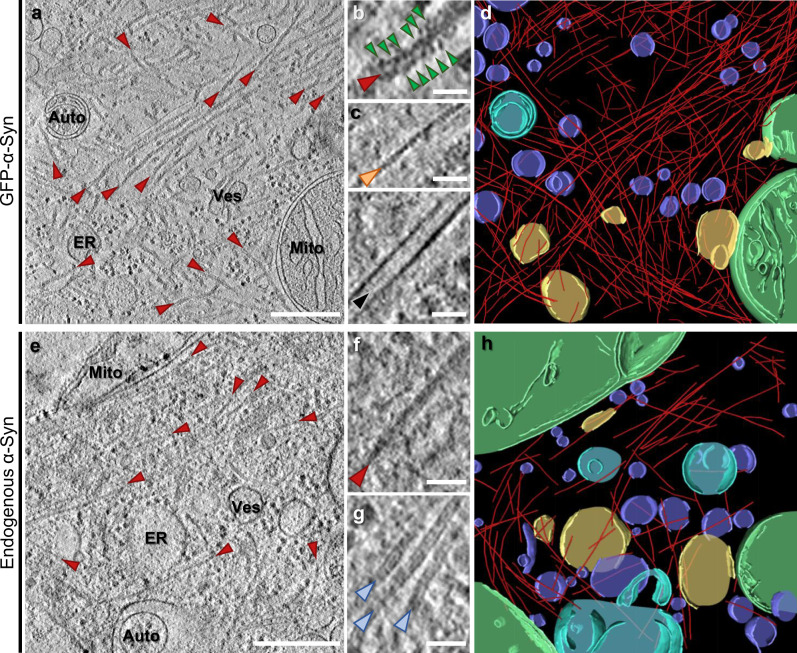
Trinkaus, V.A., Riera-Tur, I., Martínez-Sánchez, A., Bäuerlein, F.J.B., Guo, Q., Arzberger, T., Baumeister, W., Dudanova, I., Hipp, M.S., Hartl, F.U., et al. (2021). In situ architecture of neuronal α-Synuclein inclusions. Nature Communication. 12, 2110.
- 2020
-
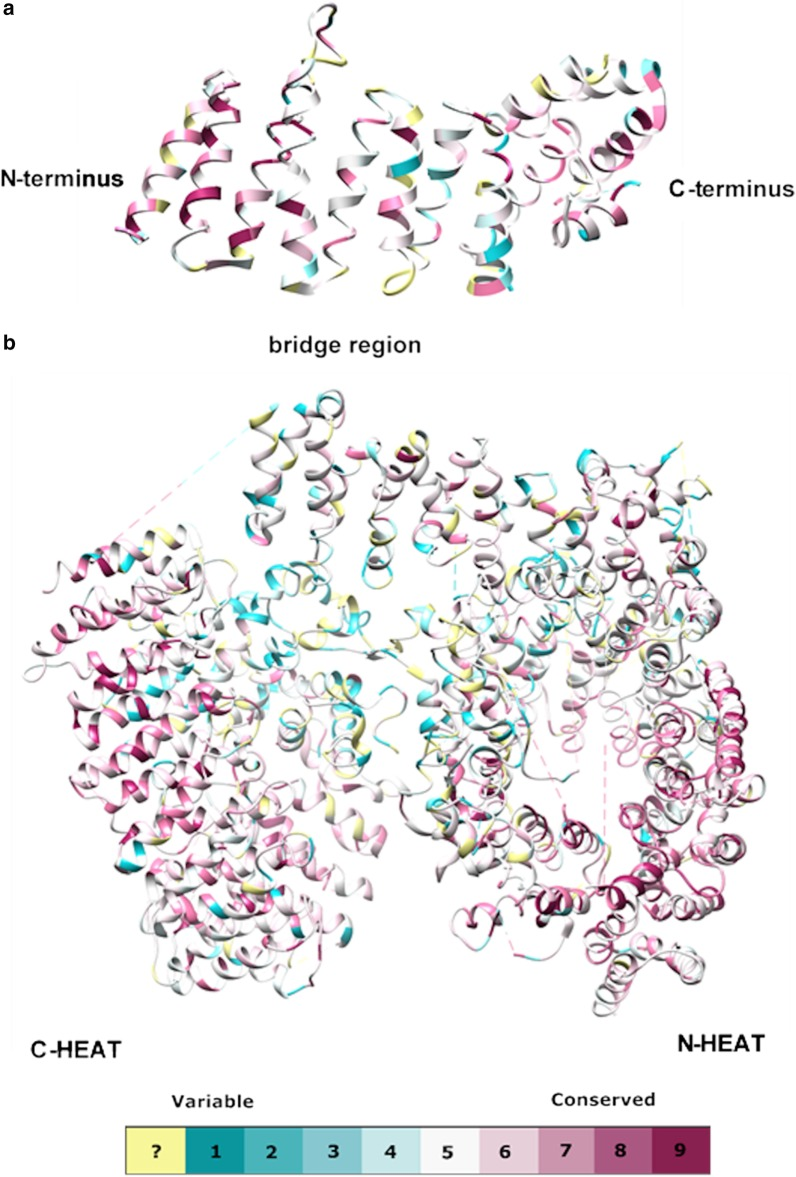
Seefelder, M., Alva, V., Huang, B., Engler, T., Baumeister, W., Guo, Q., Fernandez-Busnadiego, R., Lupas, A.N., and Kochanek, S. (2020). The evolution of the huntingtin-associated protein 40 (HAP40) in conjunction with huntingtin. BMC Evolutionary Biology 20, 162
-
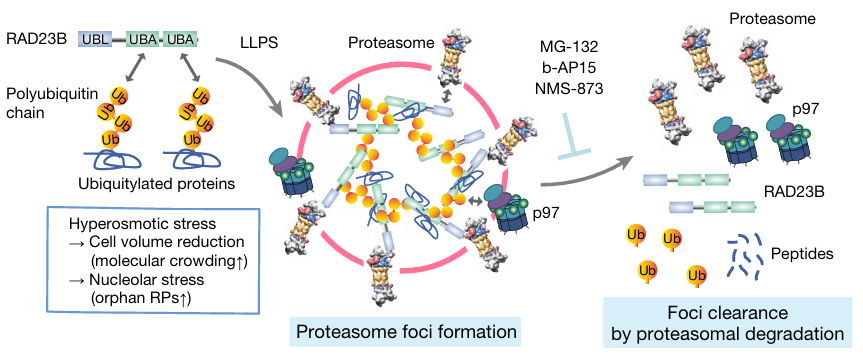
Yasuda, S., Tsuchiya, H., Kaiho, A., Guo, Q., Ikeuchi, K., Endo, A., Arai, N., Ohtake, F., Murata, S., Inada, T., et al. (2020). Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300.
- 2018
-
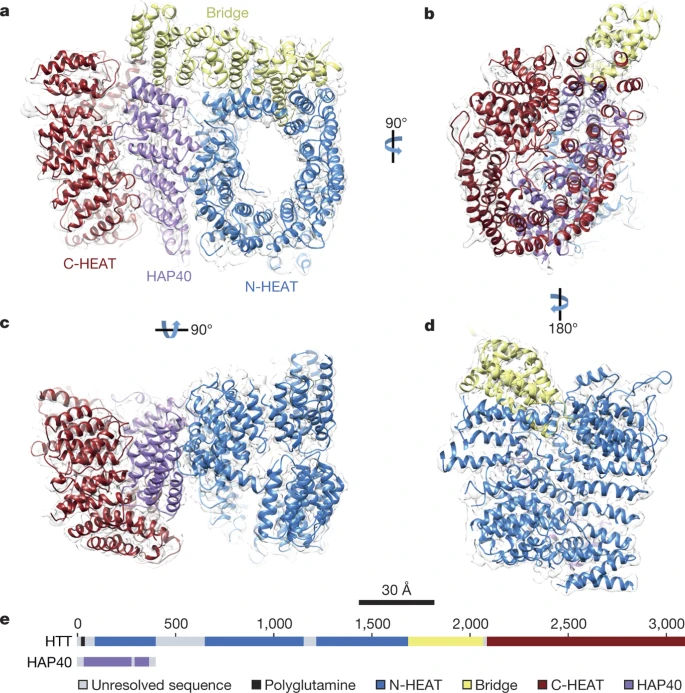
Guo, Q.*, Bin, H.*, Cheng, J., Seefelder, M., Engler, T., Pfeifer, G., Oeckl, P., Otto, M., Moser, F., Maurer, M., Pautsch, A., Baumeister, W.#, Fernandez-Busnadiego, R.#, Kochanek, S.# (2018). The cryo-electron microscopy structure of huntingtin. Nature 555, 117–120.
DOI: 10.1038/nature25502
-
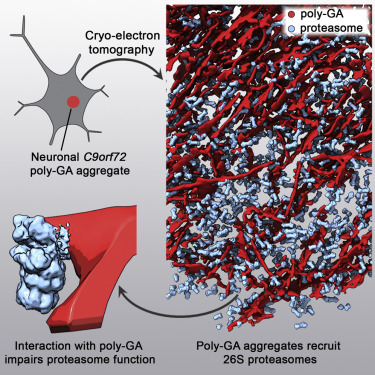
Guo, Q., Lehmer, C., Martinez-Sanchez, A., Rudack, T., Beck, F., Hartmann, H., Hipp, M.S., Hartl, F.U., Edbauer, D.#, Baumeister, W.#, Fernandez-Busnadiego, R#. (2018) In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell 172, 696-705.e612.
-
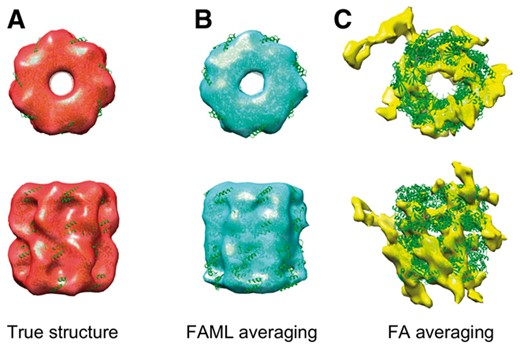
Zhao, Y., Zeng, X., Guo, Q., and Xu, M. (2018). An integration of fast alignment and maximum-likelihood methods for electron subtomogram averaging and classification. Bioinformatics 34, i227-i236.
- 2017
-
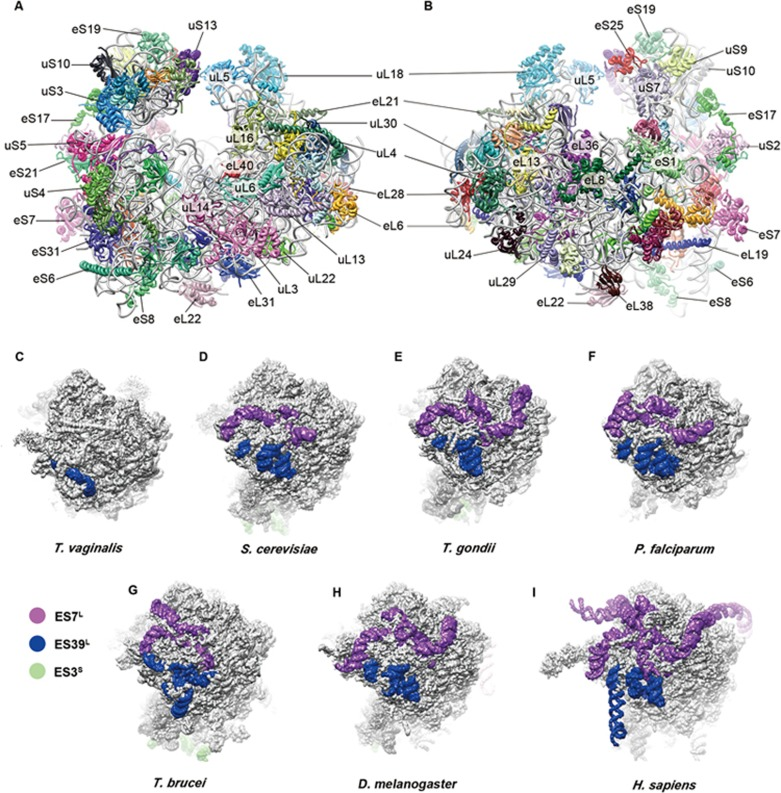
Li, Z., Guo, Q., Zheng, L., Ji, Y., Xie, Y.T., Lai, D.H., Lun, Z.R., Suo, X., and Gao, N. (2017). Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii. Cell Research 27, 1275-1288.
DOI: 10.1038/cr.2017.104
- 2015
-
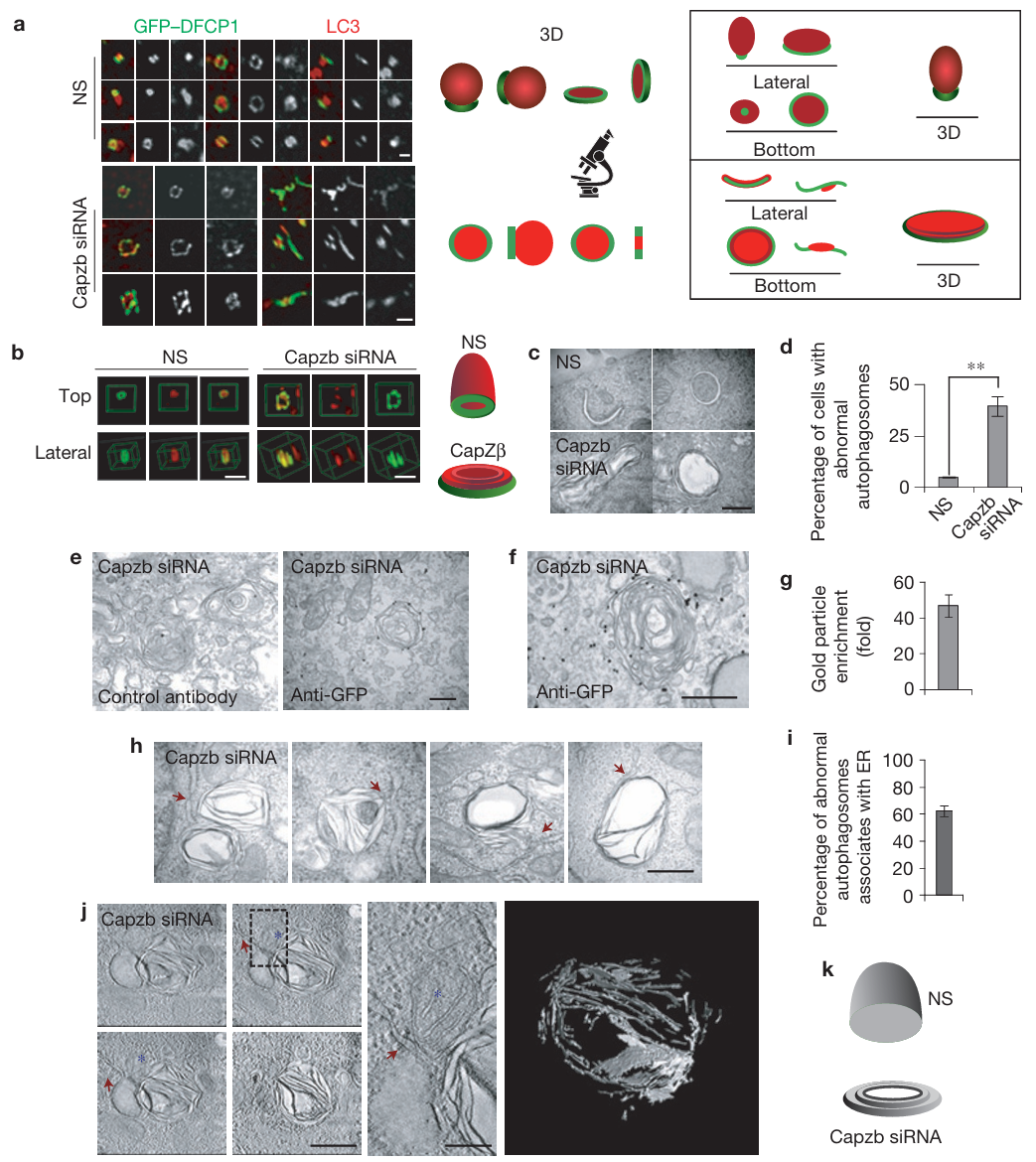
Mi, N., Chen, Y., Wang, S., Chen, M., Zhao, M., Yang, G., Ma, M., Su, Q., Luo, S., Shi, J., Xu, J., Guo, Q., et al. (2015). CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nature Cell Biology 17, 1112-1123.
DOI: 10.1038/ncb3215
- 2014
-
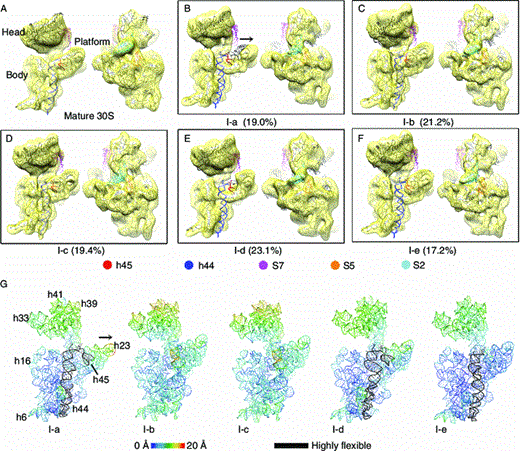
Yang, Z.*, Guo, Q.*, Goto, S., Chen, Y., Li, N., Muto, A., Himeno, H., Deng, H., Lei, J.#, and Gao, N.# (2014). Structural insights into the assembly of the 30S ribosomal subunit in vivo: functional role of S5 and location of the 17S rRNA precursor sequence. Protein Cell 5, 394-407.
-
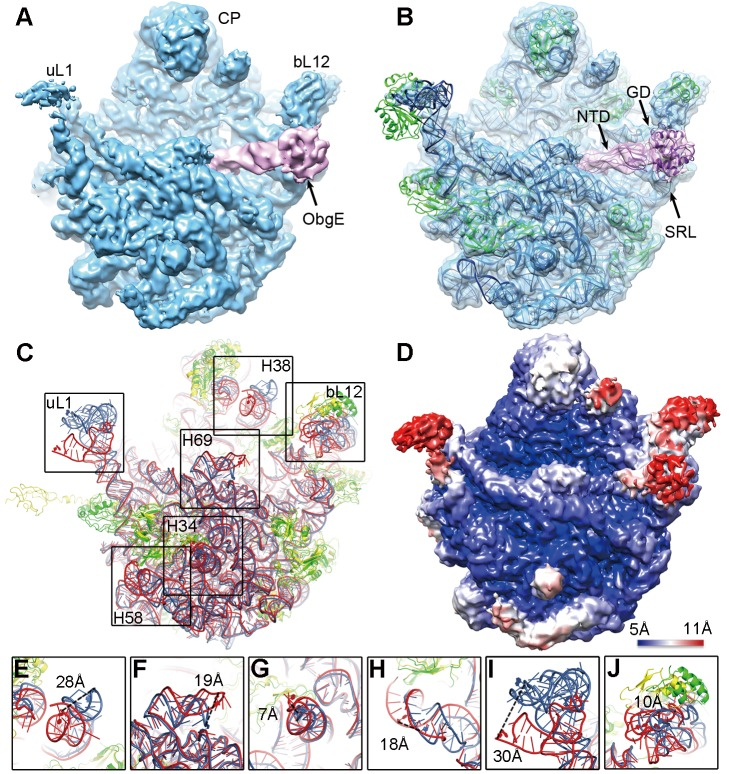
Feng, B., Mandava, CS., Guo, Q., Wang, J., Cao, W., et al. (2014) Structural and Functional Insights into the Mode of Action of a Universally Conserved Obg GTPase. PLoS Biology 12(5): e1001866.
- 2013
-
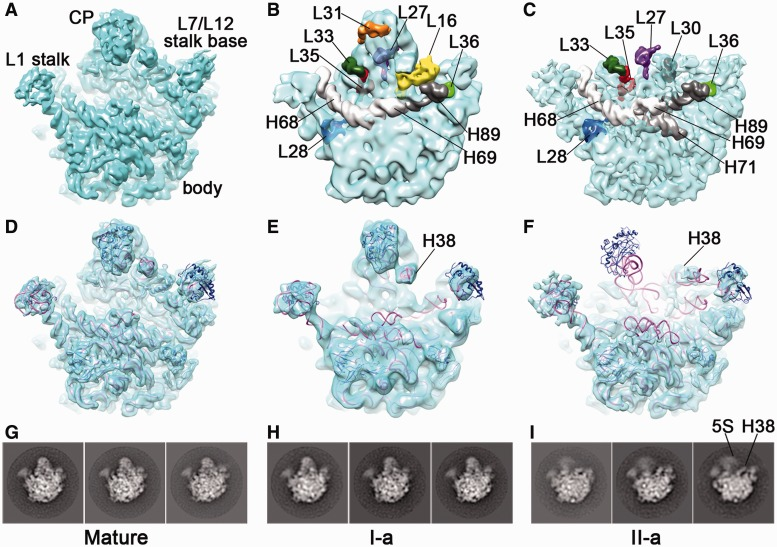
Li, N., Chen, Y., Guo, Q., Zhang, Y., Yuan, Y., Ma, C., Deng, H., Lei, J.#, and Gao, N.# (2013). Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Research 41, 7073-7083.
DOI: 10.1093/nar/gkt423
-
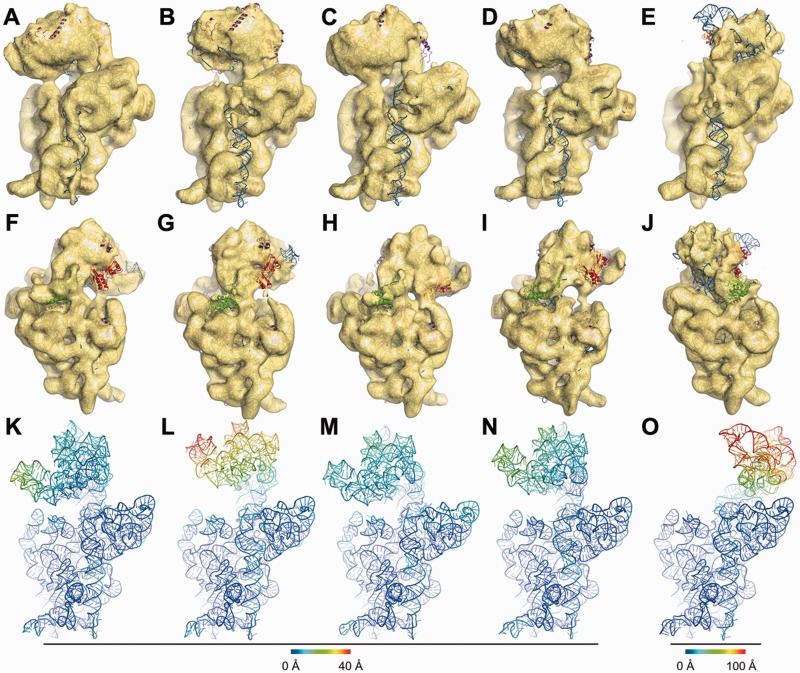
Guo, Q.*, Goto, S.*, Chen, Y.*, Feng, B., Xu, Y., Muto, A., Himeno, H., Deng, H., Lei, J., and Gao, N. (2013). Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Research 41, 2609-2620.
DOI: 10.1093/nar/gks1256
- 2012
-
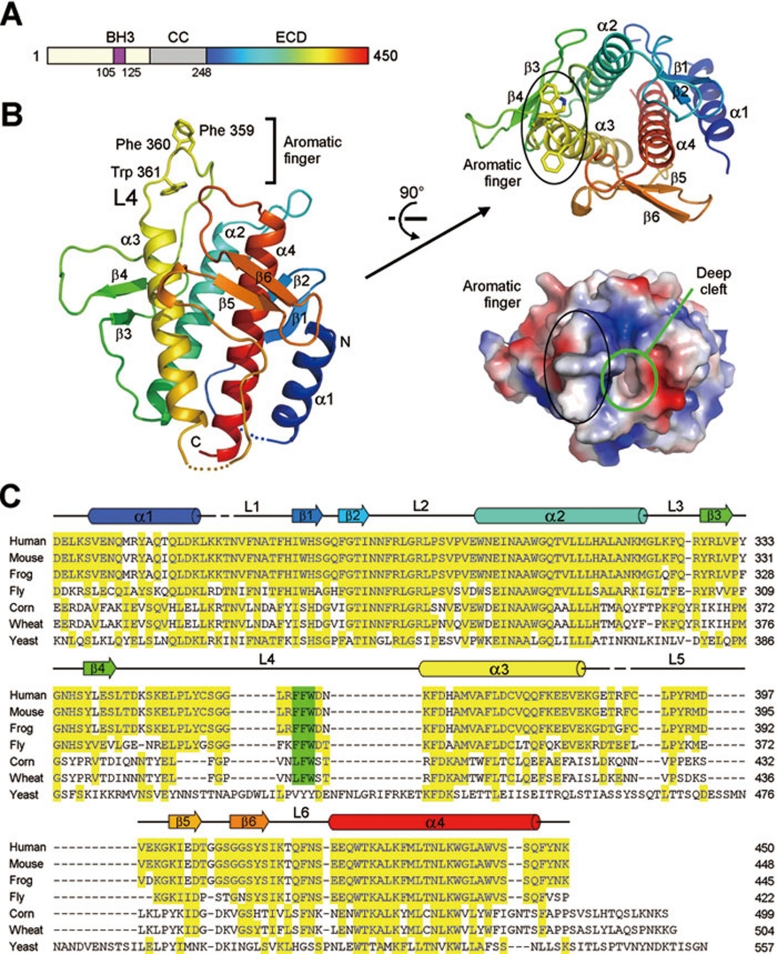
Huang, W.*, Choi, W.*, Hu, W., Mi, N., Guo, Q., Ma, M., Liu, M., Tian, Y., Lu, P., Wang, F.L., et al. (2012). Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Research 22, 473-489.
DOI: 10.1038/cr.2012.24
- 2011
-
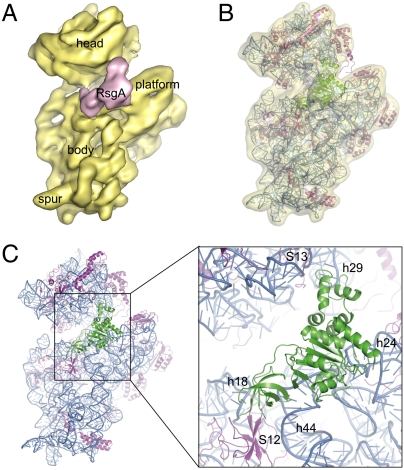
Guo, Q.*, Yuan, Y.*, Xu, Y., Feng, B., Liu, L., Chen, K., Sun, M., Yang, Z., Lei, J.#, and Gao, N.# (2011). Structural basis for the function of a small GTPase RsgA on the 30S ribosomal subunit maturation revealed by cryoelectron microscopy. PNAS 108, 13100-13105.